
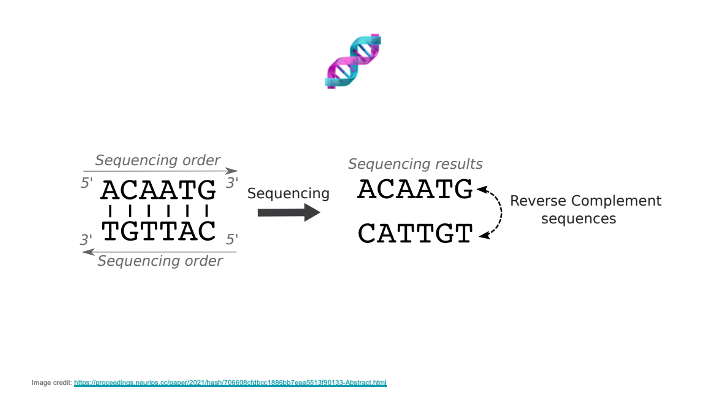
Modeling DNA introduces challenges that are distinct from those posed by natural language or proteins. First, many genomics tasks, such as predicting the effect of variants on gene expression, can entail long-range interactions, as nucleic acids even up to 1 million base pairs away from a given gene can have significant regulatory effects. Second, cellular phenotypes are often impacted by effects upstream and downstream in the genome, which requires sequence models to handle bi-directional context. Third, DNA consists of two strands that are reverse complements of each other and that carry the same information; modeling this property can significantly improve performance.
We introduce sequence modeling modules that can be applied across domains, but are specifically tailored to DNA. Specifically, we start by leveraging the recently proposed Mamba block (Gu et al. (2023)) that uses a selective state space model for long-range sequence modeling, rivaling the performance of Transformer-based models. Using this module, we develop BiMamba, a parameter/memory-efficienct, bi-directional version of Mamba. BiMamba is implemented by running a Mamba module on both a sequence and its reverse, with in and out projection weights tied. We also introduce MambaDNA, a module that extends Mamba / BiMamba to support reverse complement equivariance.
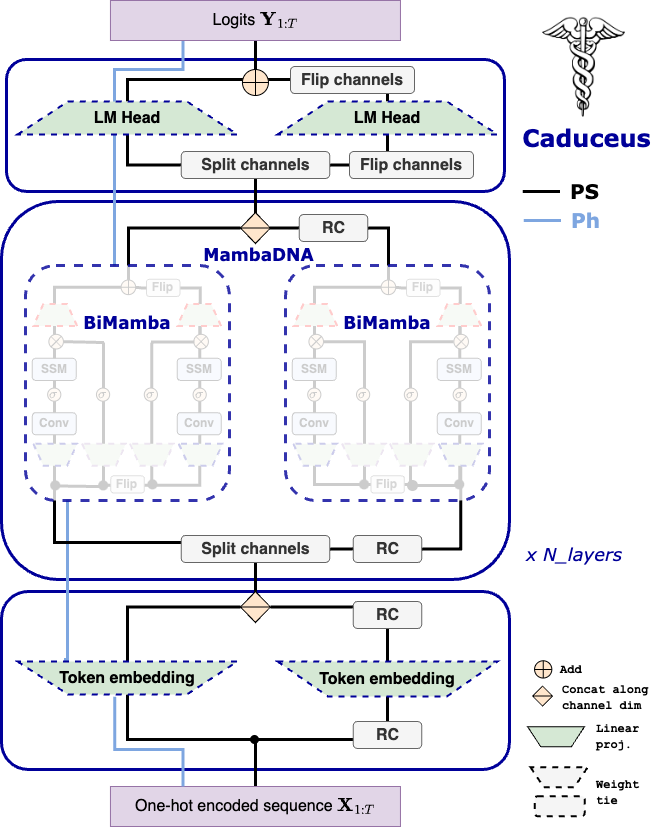
Using the sequence modeling blocks introduced above, we build Caduceus, a novel bi-directional DNA LM architecture that enforces RC equivariance. RC equivariance can be enforced in one of two ways. 1) We use the MambaDNA block in conjunction with BiMamba as the backbone of a DNA LM. With RC equivariant embedding and LM head modules, this forms Caduceus-PS (parameter sharing), the first of its kind RC equivariant LM. 2) Drawing inspiration from previous works that have investigated RC equivariant models (Zhou et al. (2022)), we also propose Caduceus-Ph (post hoc), which does not perform RC equivariant language modeling, but is rather trained with RC data augmentation and then combines predictions for forward and RC sequences post hoc at downstream task inference time.
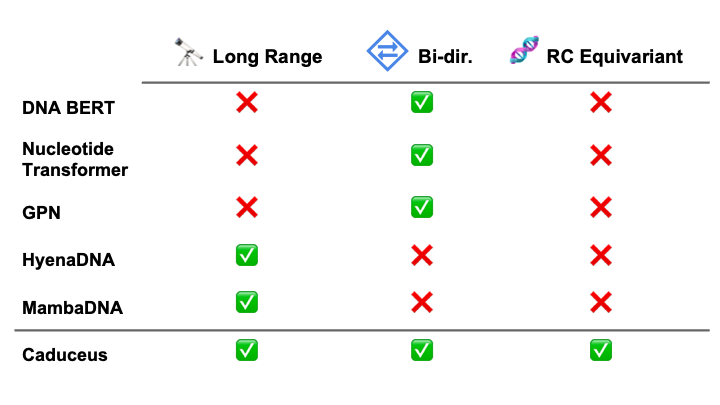
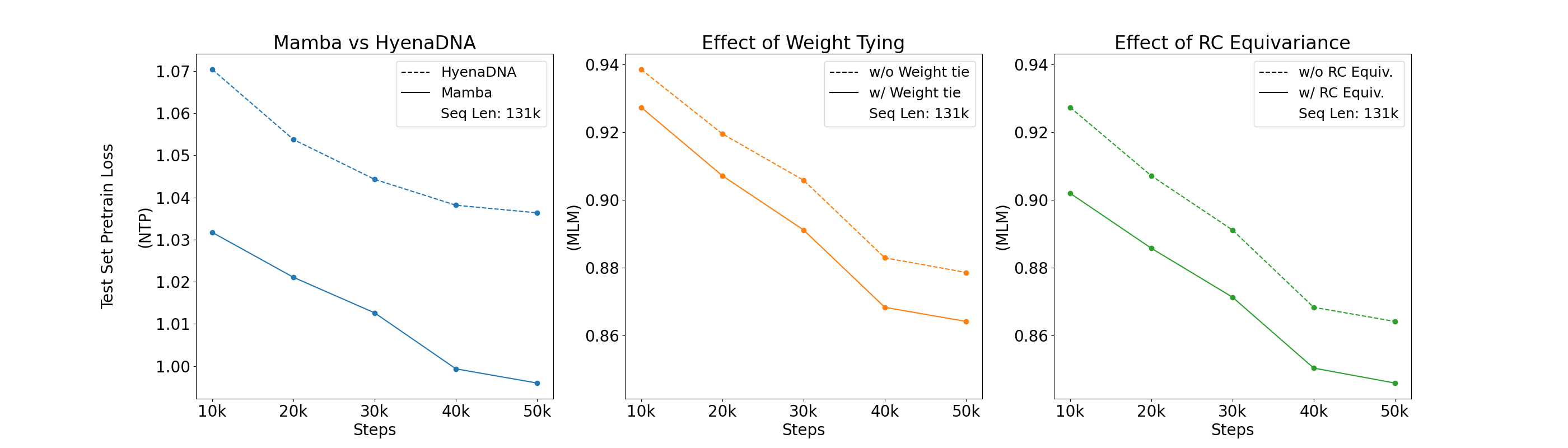
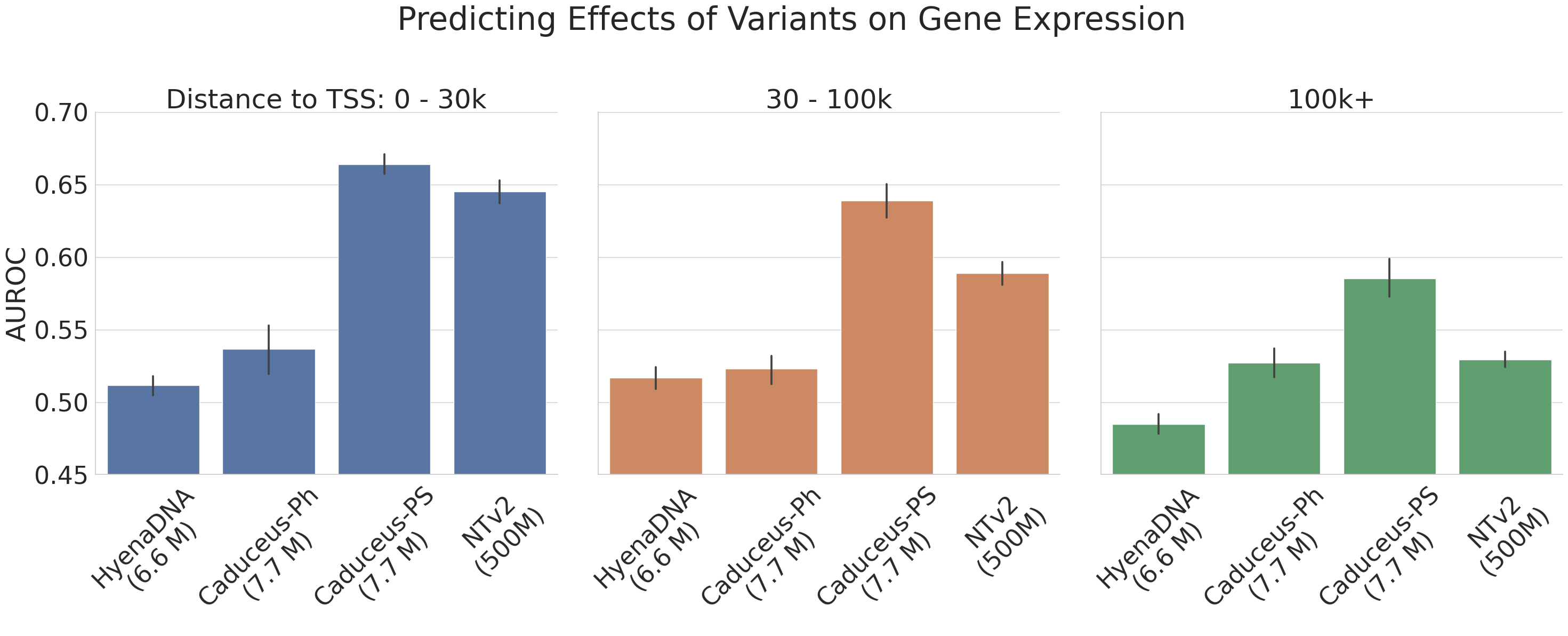
One set of benchmarks comes from the suite of tasks introduced in Nucleotide Transformer (Dalla-Torre et al. (2023)). We find that Caduceus-Ph performs competitively, even beating attention-based methods with orders of magnitude more parameters on 8 of 18 prediction tasks. Caduceus models outperform a similarly sized HyenaDNA (Nguyen et al. (2023)) model on almost all the histone marker and regulatory annotation tasks, while HyenaDNA performs better on splice site annotation.
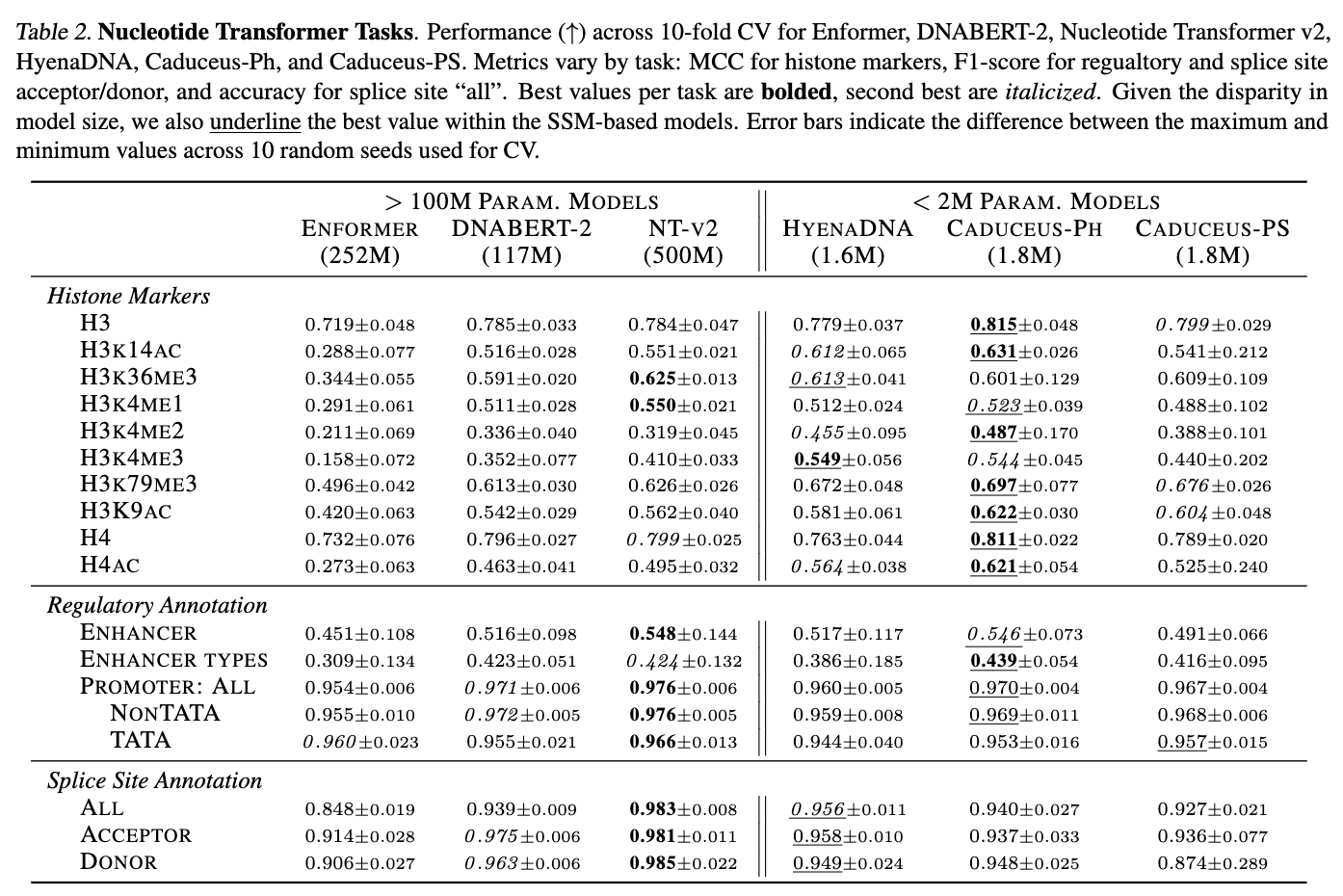
We explore the implications of long-range contexts on the task of predicting the effect of SNPs on gene expression. There is biological evidence to suggest this task indeed entails long-range interactions. Additionally it aligns well to LM pre-training objectives, which enable models to implicitly learn to recognize the effects of evolutionary pressure (e.g., conservation, co-evolution). The dataset used in this task is derived from the Enformer paper (Avsec et al. (2021)) and presented in Trop et al. (2024). From each model, we extract embeddings centered around the SNP location. We stratify the data by distance of the SNP to nearest Transcription Start Site (TSS). For each bucket, we sample 5,000 training points and fit an SVM classifier with an RBF kernel to predict VEP annotations. We report test set AUCROC mean +/- standard deviation ranges for classifiers fit on 5 random training subsets. We compare Caduceus to HyenaDNA, Nucleotide Transformer, and the suprevised baseline Enformer. As shown in the figure below the Caduceus models consistently outperform HyenaDNA, and Caduceus-PS exceeds the performance of the Nucleotide Transformer v2 (with 500M parameters), especially as distance to the nearest TSS grows. Of note, on sequences where distance to TSS exceeds 100k, Caduceus even outperforms the well-regarded Enformer baseline.
In this work, we introduced architectural innovations to the Mamba module: enabling bi-directional and RC equivariant sequence modeling. We also propose a new DNA foundation model, Caduceus, and demonstrate its ability to outperform comparably sized uni-directional Hyena-based models and Transformer-based models orders of magnitude larger in size on a range of biologically relevant tasks, most notably predicting the effect of genetic mutations on gene expression.
@article{schiff2024caduceus,
title={Caduceus: Bi-Directional Equivariant Long-Range DNA Sequence Modeling},
author={Schiff, Yair and Kao, Chia-Hsiang and Gokaslan, Aaron and Dao, Tri and Gu, Albert and Kuleshov, Volodymyr},
journal={arXiv preprint arXiv:2403.03234},
year={2024}
}